- Home
- Mission & Expertise
- Mission & Expertise
- Deep Learning
- Other applications
- Image Clinics
- Call4Help
- Support
- Software
- Scientific Collaboration
- Scientific Collaboration
- Software projects
- Teaching
- Past Activities (2018-)
- Teaching - newsletter
- Team
- Members
- Members
- Publications
- Teaching
- ScopeM staff with special abilities
- Open Positions
- Open Positions
- Social Feed
- News
- Resources
- Calendar
- Contact
- Contact
- Contact Form
Image and Data Analysis
ScopeM-ETH-Zurich, Switzerland
MISSION & EXPERTISE
Image and Data Analysis for the Life-sciences and beyond
The Image & Data Analysis group (IDA)
Part of ScopeM at ETH Zurich, Switzerland
The Scientific Center for Optical and Electron Microscopy (ScopeM) is a central scientific technology platform of ETH Zurich providing resources and services in light and electron microscopy (and related areas) as well as direct access to state-of-the-art microscopy and sample preparation equipment.
IDA specializes in the analysis of data from image-based biomedical research. This includes digital image processing, computer vision, machine learning, statistical analysis, and mathematical modeling.
What: We apply advanced image & data analysis methods to answer questions in biology and beyond.You: Someone with a question in search of a quantitative collaborator.
Us: We are a team with a multidisciplinary background in Molecular Biology, Microscopy, Computer Science, and Theoretical Physics. We actively search for interesting projects and scientific collaborations.
How: Contact us in person at an Image Clinic or write us an email. We usually respond fast and are always open to new ideas.
Deep Learning
Machine Learning for Biomedical Image Analysis

Expertise
We are happy to establish collaborations allowing scientists from different fields to apply Deep Learning techniques on their datasets. We can design & execute the whole process with you including the building of necessary annotation and the choice of tools and software.

Deep Learning for Image Segmentation
The aim of these projects is to (semi-)automatically recognize object placement and morphology (e.g. like cells and nuclei). This allows for further steps of analysis. You need to have a collection of images and annotations (typically hundreds).

Deep Learning for Image Classification
The aim of these project is to automatically provide an assignment of different images (e.g. showing cell, tissue, organism phenotypes) to specific classes (e.g. cell type, health / malignant tissue, Arabidopsis Thaliana / Lyrata, etc.). You need to have a collection of images and annotations (typically thousands).
Other applications

Single Cell Analysis
Segmentation and lineage-tracking of dividing cells
Some biological questions can only be investigated by registering behaviors of hundreds of cells over long periods of time. Such time lapse movies provide rich information, however to fully exploit this richness one needs to solve non-trivial problems such as individual cell segmentation, tracking and lineage. We have experience with analyzing this type of data in the context of various projects submerged in Systems and Synthetic Biology of Yeast (Versari et al., in preparation) and HeLa (Bertaux et al., PLoS Comp. Bio., 2014).

High-Content Screening
Image processing & analysis, statistical analysis, hit selection
We quantify, mine and make available to the users the rich information present in cellular images resulting from various screening platforms. We work on processing large datasets from high-throughput projects (10k–1m images) using mostly siRNA techniques to search for genes regulating important cellular processes e.g. in the context of infectious disease (Banerjee et al., PLoS ONE, 2013).

Advanced Microscopy Projects
Custom developed or off-the-shelf image analysis solutions
We accept assignments at nearly all levels of complexity, processing and analyzing images from diverse sources. We can help guide you through the entire process of data extraction, quantification, and statistical interpretation of results, starting from your images. We also provide advice and assistance for simpler tasks, such as preparation of professional grade figures for your publication.

Statistical & Mathematical Analysis
Time-series analysis, mathematical modelling, significance testing.
Need to get quantitative? We are comfortable in real, Fourier, wavelet, and multi-scale space. Happy to perform mathematical modelling of dynamic systems using e.g. partial differential equations. We also do generic data wrangling, statistical and time-series analysis, curve- and model-fitting, calculation of confidence intervals, significance testing and more.
Image Clinics
Bring your data or questions for a first diagnosis
Free consultations, every week
Mondays 12:00 - 14:00, telco.
Wednesdays 12:00 - 14:00, telco.
Image clinics are informal meetings held on a weekly basis open to everyone.
Aim: The purpose of these meetings is to perform a data-based brainstorming to come up with answers to your questions. We cover the three areas of image-analysis, data-analysis, and bioinformatics. Image Clinics are meant as a first contact that can later turn into a collaborative project --- sometimes we can solve your problem on the spot, but usually, we simply provide you with pointers to software and methods that will enable you to proceed in the right direction.
Booking: Booking is mandatory, please use our calendly form to secure your spot. Booking is open only for next Image Clinics (within 8 days sliding window).
Guidelines: Please follow the Call4Help spirit to present your problem: be brief, avoid jargon, and define a clear, ideal goal for Image Analysis. Please remember to bring a sample of representative data (originals, not jpg or PowerPoint slides) on an external storage device.
Sign-up: When you are at the clinics, we will fill the sign-up form, we do this to collect statistics of the most prevalent problems and departments assisted.

Call 4 Help
It's a go-to place for when you need help with your Image Data - we gather the members of Image Data Scientists' community, allowing scientists to present their problems and share their solutions.
Here you can present your case, see other presentations and share your thoughts and hints. You can also find information about upcoming Call4Help events and how to take part in them.IDA members are founders of this initiative!
SUPPORT
We provide individual consultations upon request
Experiment design
Do you want to make sure that the images you are going to register really answer the question you pose?
Together we are going to find out whether your experimental design is optimal. Together with ScopeM microscopy experts we are going to analyze the tools you plan to use, protocols of acquisition, image data processing, analysis and statistical significance.
Experiment analysis
You already have data? Do you want to make it useful for your study?
We are going to inspect together your data in the context of your scientific questions. Then we can either teach you how to process the data yourself or we can do the processing for you. Yes, it is that simple ;)
Customized image processing and analysis
Your question can not be answered with preexisting tools?
No problem, we are ready to get our hands dirty and accept the challenge of writing new tools and creating new methods to handle and process your data.
Advanced image processing and analysis
Does your question require more serious computational resources and non-trivial analysis? Would you like to have somebody do the processing and analysis for you?
We can organize processing of your data. We have human resources as well as computational and storage capabilities. We can set up your project within the ScopeM infrastructure.
Image Ethics
Paraphrasing and quoting from:
Cromey, D. W. (2010). Avoiding Twisted Pixels: Ethical Guidelines for the Appropriate Use and Manipulation of Scientific Digital Images. Science and Engineering Ethics, 16(4), 639–667.
Image processing is a form of communication.
Image processing is not a form of artistic expression.
Image processing is the mathematical manipulation of the underlying numbers in a digital image.
Scientific Collaborations
How to start a collaborative project with us

Individual Projects
By appointment, HPZ G31, ETH Zurich Hönggerberg
We are always willing to discuss new projects and approach these as scientific collaborations.
Individual projects are often extensions of a first contact at the Image Clinic.
If your task is more complex and we together decide to continue working on it, an Individual Project provides us with the means to proceed. Before initiation, we will agree on a specification, estimation of required time and delivery dates.
What can we do for you?
Our expertise span many domains: we can help you with experimental design and planning, raw image data analysis, statistical data analysis, and of course with publications --- providing the part of the manuscript that covers what we did, setting up data or code repositories and more. We also support grant writing: Our expertise is not limited to image data analysis but includes most potential data sources.
The project work can typically be sub-divided as follows:
- First Exploration - mandatory first step in any project, booked as the equivalent of 15 hrs. A report is delivered.
- Project work - as agreed with the user after the initial report
- Follow-up - due to change in user-request or directly a result of new insights. Only upon prior agreement with the user.

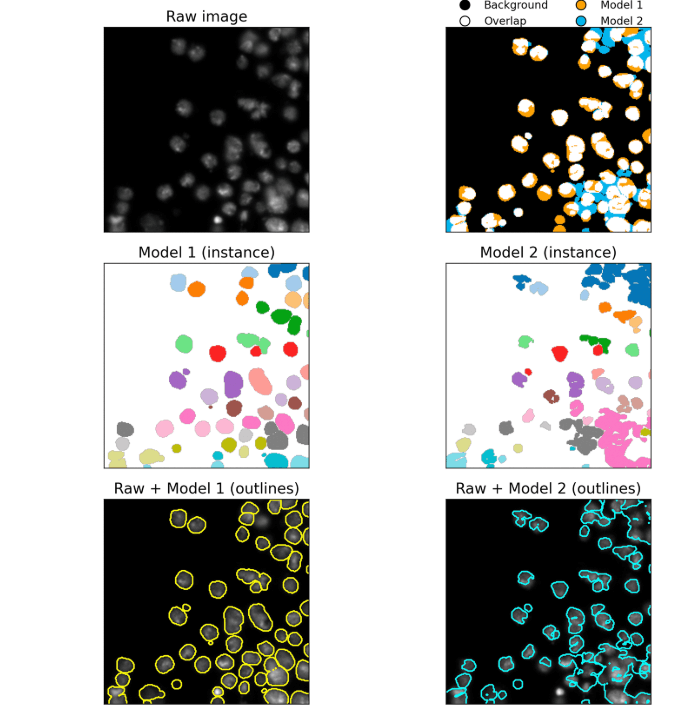
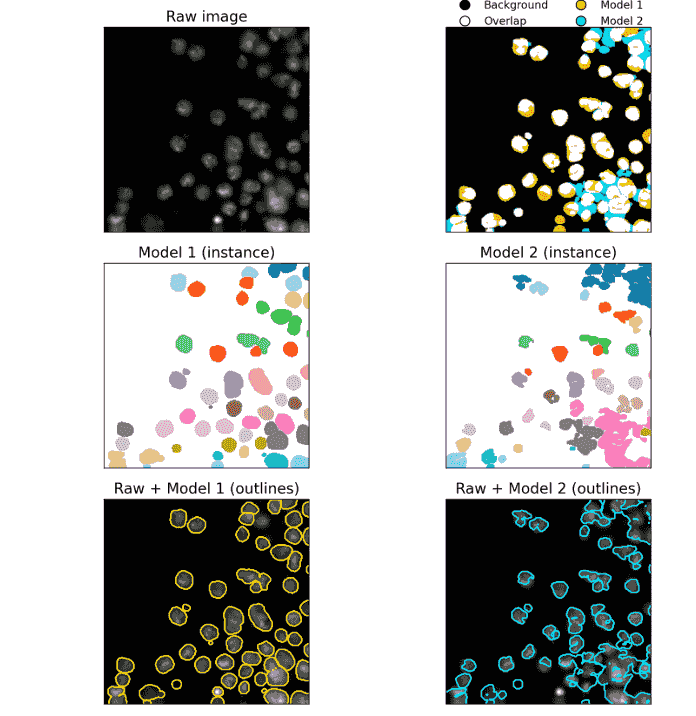
BISCUIT
BISCUIT (BioImage Segmentation Comparison Utility and Interactive Tool) is an open-source platform for visually comparing state-of-the-art cell segmentation models on microscopy images.

Essential Vision - Hololens
An innovative application dedicated for Microsoft HoloLens headset – is a new tool for visualization of time series 3D models and their simulation data.
Partners: ETH Zurich
cellstar-algorithm.org
Automatic tracking of cells in time-lapse microscopy is required to investigate a multitude of biological questions. To limit manipulations during cell line preparation and phototoxicity during imaging, brightfield imaging is often considered. Since the segmentation and tracking of cells in brightfield images is considered to be a difficult and complex task, a number of software solutions have been already developed.
CellStar is one of such algorithms. It is optimized to segment and track images of budding yeast cells growing in monolayer (e.g. images from microfluidic chambers), however the algorithm can be also used to track other round objects (in brightfield as well as fluorescent images).
Partners: ETH Zurich, INRIA, University of Lille, CNRS, University of Wroclaw, Broads Institute (MIT/Harvard)

taxonify.org
Taxonify is a platform that expedites the process of species annotation for automated classification. It supports image browsing and annotation in a way that is optimal for taxonomists and their annotation workflow. Taxonify is free and open source - you can download, modify it and use it according to your needs!
It's currently used in the project Aquascope for annotation of phytoplankton and zooplankton images coming from automated underwater imaging cameras in lake Greinfensee (Switzerland). The software was funded by the Swiss National Science Foundation project 310030L_182124 (Aquascope). You can check out more about Aquascope project on their website - www.aquascope.eawag.chPartners: ETH Zurich, EAWAG
Autotube
AutoTube quantifies vascular parameters such as the vessel area, width, skeleton length or the number of branching points of vascular networks in tissues or in invitro assays. AutoTube is freely available, comprises an intuitive Graphical User Interface (GUI) and helps to perform otherwise highly time-consuming image analyses in a rapid, automated objective and reproducible manner.
Partners: ETH Zurich
Teaching
Image Analysis courses we organise or teach in

Introduction for Image Analysis using ImageJ / FIJI
Date: 05-06th March, 2026
Place: ETH Hönggerberg, HPZ E35, Zurich
Application: https://forms.gle/r956jrr9nKsyz5kU7
Costs: 120 CHF Academia / 200 CHF Industry; charged via PPMS
The training runs over two consecutive afternoons (13h-18h) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands-on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH and UZH members or external users upon prior agreement.
Course credit: 1 ECTS, for PhD students.
Introduction to image analysis with IMARIS
23-24th March 2026
Irchel campus
Application: https://forms.cloud.microsoft/e/ZAKABSAXNk (deadline 2nd March 2026)
Join us for a hands-on training on Imaris 11, led by an expert from Oxford Instruments and coorganized together with ZMB. This workshop is designed for UZH & ETH and ScopeM/ZMB users interested in microscopy image analysis.
The new Imaris 11 introduces a redesigned interface and powerful experimental pipelines for streamlined, automated analysis workflows, including one-click export and batch processing.
Participants will work directly with the software on virtual machines and can also analyze their own data with guidance from the instructor. Topics include 3D segmentation, object tracking, scientific visualization, and 3D annotation.
SwitZerland's Image and Data Analysis School 2026
5-10th July 2026, Lausanne
Application opens later in 2026
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
MORE: 2026.zidas.org
Past Activities (2014 - 2026)

Zurich Winter School 2026 - Practical Course in Advanced Microscopy
January 2026
Application closed
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique.
Theoretical sessions will cover basic and advanced knowledge about all the different techniques and topics. Practical work consists of five different modules each covering a specific topic:
• Advanced Light Microscopy
• Super-Resolution Light Microscopy
• Sample Preparation for Electron Microscopy
• 3D Correlative Light and Electron Microscopy
• Volume Scanning Electron Microscopy
Introduction to image analysis with IMARIS
12-13 November 2026
Application: https://forms.gle/XPgtovkt52PMhGjU6 (deadline 3rd Nov. 2025)
Join us for a hands-on training on Imaris 11, led by an expert from Oxford Instruments. This workshop is designed for ETH Zurich and ScopeM users interested in microscopy image analysis.
The new Imaris 11 introduces a redesigned interface and powerful experimental pipelines for streamlined, automated analysis workflows, including one-click export and batch processing.
Participants will work directly with the software on virtual machines and can also analyze their own data with guidance from the instructor. Topics include 3D segmentation, object tracking, scientific visualization, and 3D annotation.

SwitZerland's Image and Data Analysis School 2025
6-11th July 2025
Application closed.
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
MORE: 2025.zidas.org

Zurich Winter School - Practical Course in Advanced Microscopy
20-24 January 2025
Application deadline: Sunday, 17th of November 2024
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique.
Theoretical sessions will cover basic and advanced knowledge about all the different techniques and topics. Practical work consists of five different modules each covering a specific topic:
• Advanced Light Microscopy
• Super-Resolution Light Microscopy
• Sample Preparation for Electron Microscopy
• 3D Correlative Light and Electron Microscopy
• Volume Scanning Electron Microscopy
MORE: Webpage

Zurich Winter School - Practical Course in Advanced Microscopy
Date: 9-10th October, 2025
Place: ETH Hönggerberg, Zurich
Costs: 120 CHF Academia / 200 CHF Industry; charged via PPMS
The training runs over two consecutive afternoons (13h-18h) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands-on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH and UZH members or external users upon prior agreement.
Course credit: 1 ECTS, for PhD students.

Introduction for Image Analysis using ImageJ / FIJI
Date: 12-13th September, 2024
Place: ETH Hönggerberg, Zurich
Application link (deadline 2024-09-06)
Costs: 120 CHF Academia / 200 CHF Industry; charged via PPMS
The training runs over two consecutive afternoons (13h-18h) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands-on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH and UZH members or external users upon prior agreement.
Course credit: 1 ECTS, for PhD students.

SwitZerland's Image and Data Analysis School 2024
23-28th June 2024
Application link (deadline 2024-03-17)
600 EUR (academia) / 1800 EUR (industry)
Place: Zurich, Switzerland
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
MORE: 2024.zidas.org

SwitZerland's Image and Data Analysis School 2023
16-11 of July 2023
Place: Lausanne, SwitzerlandDate: 7-9 October 2019
Application: Through ETH's PSC PhD programme
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
If you answered yes to some of the above, then this school is for you!
MORE: 2023.zidas.org

17th EXCITE Zurich Summer School on Biomedical ImagingFrontiers in Plant Sciences: Best practice in rendering digital images for publication
September 2023
Place: Zurich, Switzerland
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week.
MORE: https://excite.ethz.ch/education/summer-school.html
Course Objectives
This course aims to focus on a key step in the scientific process: the rendering of scientific data in a form that can be shared with colleagues – usually in the form of a published research paper, a preprint or a scientific talk. This step is critical as it is all too easy to misrepresent ones research findings in the rush to publish and given the pressures to publish ‘high impact’ research papers. The course aims to complement other training at the PSC PhD programme and therefore focuses on image based data and digital processing of such data for publication.
Course Program
Topics covered will include:
- Best practice and responsible conduct when acquiring and processing image based data. How to assemble a compelling paper that nevertheless represents the scientific findings in an accurate, unbiased manner.
- Reproducibility: critical evaluation of the ‘reproducibility crisis’ and how to render papers more reproducible. New policies and mechanisms that enhance reproducibility.
- The most frequent digital image aberrations observed in the literature and how to prevent them.
- What to do if you find image aberrations in unpublished work and published work (including your own data, data from your lab and other labs).
- How to peer review image based data and screening mechanisms at journals for digital image aberrations.
- How journals deal with image aberrations – emerging best practice.
Number of Participants: 16
Credit Points: 1 ECTS

Introduction for Image Analysis using ImageJ / FIJI
Date: 13-15th of March 2023 (afternoons: 1-6 PM)
Place: ETH Hoenggerberg
The training runs over three consecutive afternoons (1 PM - 6 PM) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands-on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH and UZH members or external users upon prior agreement.
Course credit: 1 ECTS, for PhD students.

16th EXCITE Zurich Summer School on Biomedical Imaging
Date: September 2022
Place: Zurich
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week. WEBSITE

SwitZerland's Image and Data Analysis School 2022
Date: July 2022
Place: Lausanne
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
MORE: 2022.zidas.org
Deep Learning for Image Analysis
EMBL Course
Date: January 2022
Place: VIRTUAL
This is a blended learning course. We will begin with 3 pre-course sessions (one in December 2021, two in January 2022) which will bring everyone to a good starting position for a week-long intensive virtual course in February 2022. The 3 pre-course sessions will have associated homework. The week-long course in February 2022 will include a few talks, but will mostly be devoted to hands-on work on real data, in small groups of 3-4 participants.

Deep Learning for Image Analysis
EMBL Course
Date: 8 - 12 Feb 2021
Place: VIRTUAL
This is a blended learning course. We will begin with 3 pre-course sessions (one in December 2020, two in January 2021) which will bring everyone to a good starting position for a week-long intensive virtual course in February 2021. The 3 pre-course sessions will have associated homework. The week-long course in February 2021 will include a few talks, but will mostly be devoted to hands-on work on real data, in small groups of 3-4 participants.

Date: 1st September - 12th September 2021
Place: Zurich
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week. WEBSITE

SwitZerland's Image and Data Analysis School 2021
Date: 27th September - 1st October 2021
Place: VIRTUAL
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
Is this school for you?
- Are you a life-science researcher with a pressing need to quantify your light-microscopy images?
- Are you uncertain about how to: Best calculate co-localisation, do deconvolution, automate the counting of cells, track objects over time, handle massive amounts of image data, record your image-analysis workflows in a reproducible manner?
- If you answered yes to some of the above, then this school is for you!
MORE: 2021.zidas.org
Date: 28th June - 2nd July 2021
Place: Virtual
This course is aimed at early-career scientists as well as staff scientists working in microscopy or image analysis facilities who already have experience in bioimage analysis. Moreover, participants should already provide or show the clear intent of providing bioimage analysis support to other researchers with less or no experience in bioimage analysis.
- Microscopy image quality control and image restoration
- Advanced image segmentation and complex cell phenotyping
- Handling large microscopy images (such as whole slide scans) and big N-dimensional data
- Neural-networks for image restoration, segmentation, and object classification.
- Co-localisation and spatial statistics
- Train the trainer: how to teach image analysis

14th EXCITE Zurich Summer School on Biomedical Imaging
Zurich, 31 August - 11 September 2020
Application: Here
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week.

SwitZerland's Image and Data Analysis School (ZIDAS 2020)
29 June to 3 July 2020, EPFL, Lausanne, Switzerland [VIRTUAL]
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
MOOC
Before the start of the course, you’re expected to validate the IPA4LS on-line course . You’ll have some videos, exercises and quizzes to gauge your IDA knowledge. We ask you to follow the “extra week” dealing with the ImageJ Macro language.
Learning objectives
You will learn the fundamentals of image analysis, including basic macro programming in ImageJ/Fiji as well as other software solutions.
In the first part of the week we will also cover the process of image-formationas it pertains to image analysis: Resolution, correct exposure, point-spread functions, detector noise, Shannon's sampling theorem, and aliasing. All with a clear focus on application in the lab.
In the second half of the week there will be a number of focused topics, building on what was learned during the first days, e.g.:
- Deep Learning for image restoration and segmentation.
- Co-localisation, i.e., spatial correlation analysis and statistics.
- Data Analysis using KNIME.
- De-convolution of microscopy images.
- Tracking of particles and cells in time-lapse recordings.
- Stitching and registration of stacks of large image data.
- Hands-on training with image-analysis software solutions such as ilastik, CellProfiler and QuPath.
For details, go here: 2020.zidas.org

The 4th NEUBIAS Conference & Symposium*
The 4th Annual Conference of NEUBIAS, gathering the whole BioImage Analysis Community into a multi-faceted event, offers two parallel Training Schools for selected early career and advanced analysts and a Working Meeting (Taggathon) for invited taggers, which precede a Large Symposium dedicated to recent Scientific Developments and Open Tools in BioImage Analysis. Participation in the Symposium is open to all from academia and industry, where participants will be accepted until all spots are filled.
Training Schools and Taggathon are taking place at the Interdisciplinary Institute for Neurosciences of the University of Bordeaux from February 29th to March 3rd, 2020, followed by the symposium which is hosted at the ENSEIRB MATMECA campus in the city center of Bordeaux on March 4, 5 and 6, 2020.

20 - 24 Jan 2020, EMBL Heidelberg, Germany
20 - 24 Jan 2020, EMBL Heidelberg, Germany
This is a blended learning course on Deep Learning for Image Analysis, consisting of 3 online sessions with associated hands-on exercises and a week-long onsite session at EMBL Heidelberg. This course is aimed at both core facility staff and research scientists. Prerequisites for this workshop are programming skills in Python and ideally Tensorflow, Keras or Pytorch as well as basic knowledge of machine learning theory.
Participants should provide an outline of one image analysis task they would like to work on during the on-site part of the course. Neural networks have been successfully applied to various medical and biological imaging modalities including PALM/STORM, light sheet fluorescence microscopy, high-throughput microscopy, electron microscopy, X-ray tomography. However, they require observation-outcome-pairs for training. Ideally, you will be provide annotated images for network training during the course. After this course you should be able to:- Explain the fundamentals of machine learning methods suitable for image analysis
- Consult users/colleagues in strategies to obtain ground truth
- Give advice in training and using a CNN for image analysis
- Perform simple quality control on the results

Frontiers in Plant Sciences: Best practice in rendering digital images for publication
Date: 7-9 October 2019
Application: Through ETH's PSC PhD programme
This is a blended learning course on Deep Learning for Image Analysis, consisting of 3 online sessions with associated hands-on exercises and a week-long onsite session at EMBL Heidelberg. This course is aimed at both core facility staff and research scientists. Prerequisites for this workshop are programming skills in Python and ideally Tensorflow, Keras or Pytorch as well as basic knowledge of machine learning theory.
Participants should provide an outline of one image analysis task they would like to work on during the on-site part of the course. Neural networks have been successfully applied to various medical and biological imaging modalities including PALM/STORM, light sheet fluorescence microscopy, high-throughput microscopy, electron microscopy, X-ray tomography. However, they require observation-outcome-pairs for training. Ideally, you will be provide annotated images for network training during the course. After this course you should be able to:- Explain the fundamentals of machine learning methods suitable for image analysis
- Consult users/colleagues in strategies to obtain ground truth
- Give advice in training and using a CNN for image analysis
- Perform simple quality control on the results

Zurich Image and Data Analysis School 2019 (ZIDAS 2019)
18-23 June 2019. Zurich, Switzerland
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
You will learn the fundamentals of image analysis, including basic macro programming in ImageJ/Fiji as well as other software solutions.
In the first part of the week we will also cover the process of image-formation as it pertains to image analysis: Resolution, correct exposure, point-spread functions, detector noise, Shannon's sampling theorem, and aliasing. All with a clear focus on application in the lab.
In the second half of the week there will be a number of focused topics, building on what was learned during the first days, e.g.:
- Deep Learning for image restoration and segmentation
- SMLM (Single-Molecule Localisation Microscopy) data analysis
- Hands-on training with image-analysis software solutions such as ilastik and CellProfiler
For details, go here: 2019.zidas.org

Introduction to Image Analysis using FIJI/ImageJ
24th-26th April 2019
Application: CLOSED
CLOSED
12-24.01.2014, ETH Zurich Hönggerberg
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique.

Zurich Winter School - Practical Course in Advanced Microscopy
20th to 25th of January 2019.
ETH Zurich and the University of Zurich
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique. Students will be able to apply this technique to their own present and future projects. Practical work consists of six different modules covering a specific topic each (please see description below). Each practical module lasts throughout the whole course. Students will participate in one module only and use state-of-the-art instrumentation. Theoretical joined sessions will in turn cover basic knowledge about all the different techniques for all students.
The course is jointly organised by EXCITE, the Scientific Center for Optical and Electron Microscopy (ScopeM, ETHZ) and the Center for Microscopy and Image Analysis(ZMB, University of Zurich).

Machine Learning for Image Analysis
29th to 31st October 2018. EMBL Heidelberg
This is a blended learning course on Machine Learning for Image Analysis, consisting of three online sessions with associated hands-on exercises prior to the workshop, a three day face-to-face workshop at EMBL Heidelberg and two optional online sessions with associated hands-on exercises after the workshop. The course is jointly organised by CORBEL, EMBL, German BioImaging and NEUBIAS (COST Action CA15124).

12th Zurich Summer School on Multiscale Biomedical Imaging
3-14 September 2018, Zurich
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week.

Zurich Image and Data Analysis School 2018 (ZIDAS 2018)
19-24 June 2018, Zurich, Switzerland
CLOSED
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
You will learn the fundamentals of image analysis, including basic macro programming in ImageJ/Fiji as well as other software solutions.
In the first part of the week we will also cover the process of image-formation as it pertains to image analysis: Resolution, correct exposure, point-spread functions, detector noise, Shannon's sampling theorem, and aliasing. three-day with a clear focus on application in the lab.
In the second half of the week there will be a number of focused topics, building on what was learnt during the first days:
- Scripting in MATLAB
- Tracking particles and cells in time-lapse recordings
- De-convolution of microscopy images
- Stitching and registration of stacks of large image data

2nd NEUBIAS conference
27th Jan. to 2nd Feb. 2018 Hungary
NEUBIAS is a workshop & a conference to exchange the newest findings, applications, and cutting-edge developments in Bioimage Analysis, machine learning, data mining, and storage. European Bioimage Analysts, an emergent group within the bioimaging analysis community, organize this event, bringing together an international, interdisciplinary community of scientists in life and computer sciences.

Zurich Winter School - Practical Course in Advanced Microscopy
21-26 January 2018, Zurich
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique. Students will be able to apply this technique to their own present and future projects. Practical work consists of six different modules covering a specific topic each (please see description below). Each practical module lasts throughout the whole course. Students will participate in one module only and use state-of-the-art instrumentation. Theoretical joined sessions will in turn cover basic knowledge about all the different techniques for all students.

Introduction to Digital Pathology using QuPath
21-23.11.2017 ETH and UZH
Application: Deadline 12-noon, October 6th, 2017 (prolongated deadline)
This course is an introduction to Image Processing and Analysis using QuPath, an open source software dedicated to processing histopathological datasets. The course will be lectured by Pete Bankhead, a lead developer of QuPath, and an author of an excellent ImageJ book, supported by ScopeM IDA: Szymon Stoma and Simon F. Nørrelykke. Pete recently left Queen's University Belfast to reinforce Phillips Healthcare Unit led by Peter Hamilton. The attendees will learn the fundamentals of image analysis including how to do basic Macro programming in QuPath for automated batch analysis of images, and will be introduced to visualisation and explorative data analysis after extraction of numerical data from images.

Zurich Image and Data Analysis School 2017
16-20.10.2017 ETH and UZH
Application: CLOSED
This one-week school provides a hands-on introduction to image processing and analysis, with an emphasis on biologically relevant examples.
You will learn the fundamentals of image analysis, including basic macro programming in ImageJ/Fiji as well as other software solutions.
In the first part of the week we will also cover the process of image-formation as it pertains to image analysis: Resolution, correct exposure, point-spread functions, detector noise, Shannon's sampling theorem, and aliasing. All with a clear focus on application in the lab.
In the second half of the week there will be a number of focused topics, building on what was learnt during the first days:
- Scripting in MATLAB
- Tracking particles and cells in time-lapse recordings
- De-convolution of microscopy images
- Stitching and registration of stacks of large image data

11th Zurich Summer School on Multiscale Biomedical Imaging
4-15.09.2017 ETH and UZH
Application: CLOSED
The EXCITE Summer School on Biomedical Imaging is dedicated to teaching the basics of biomedical imaging alongside an overview of applications which are vital to understand recent advances and current challenges in biological and medical imaging. A range of cutting-edge techniques and analysis tools are discussed. Leading scientists present lectures and hold practical sessions. Opportunities to network with industry are provided. The school takes two weeks and awards 4 ECTS upon passing a written exam at the end of the second week.
Excellent MSc and PhD students as well as scientists having a background in life sciences, biology, computer science, engineering, mathematics or physics are invited to apply.

CFIM Image Analysis Course 2017
19-23.06.2017, Copenhagen
Application: CLOSED
This course is an introduction to image processing and analysis, with a focus on biologically relevant examples. The attendees will learn the fundamentals of image analysis including how to do basic Macro programming in Fiji (ImageJ) for automated batch analysis of images, use different software solutions for image analysis, and will be introduced to visualisation and explorative data analysis after extraction of numerical data from images.

BIAS2017: Bio-Image Data Analysis 2017
(EMBL Master Course)
14 - 20 May 2017, EMBL Heidelberg, Germany
Application: CLOSED
12-24.01.2014, ETH Zurich Hönggerberg
This course will focus on computational methods for analysing images of single molecules, cells and tissues, to boost the learning process of participants who have an immediate need to deploy image analysis in their own research. For this, the course extends from basic principles to the actual implementation of workflows using scripting. By increasing their image analysis literacy, participants will greatly enhance the scope, creativity and achievements of their research projects. Expert knowledge will be gathered to create a world-leading course for image analysis in the field of biology.

Introduction to Image Analysis using FIJI/ImageJ
The training runs over four consecutive afternoons (1 PM - 6 PM) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands-on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH and UZH members or external users upon prior agreement. A 100 CHF fee (400CHF for the industry) is used for printed materials and coffee during breaks. Please apply via an online application.
Course credit: 1 ECTS, for PhD students.
Bioimage Analysis Training Schools
12-17.03.2017 Lisbon, Portugal
NEUBIAS: Network of European Bioimage Analysts
This NeuBIAS training school series is based on the highly successful BIAS courses (@ EMBL) and EuBIAS symposia courses, and will be provided by NeuBIAS faculty and new invited trainers. The program will include computational methods and tools for analysing images of molecules, cells and tissues, tailored for Staff Scientists working in, or in association with, microscopy/bioimaging facilities, who can then provide support and training for researchers who have an immediate need to deploy image analysis in their research. For this, the course extends from basic principles to the actual implementation of workflows using scripting.
Introduction to Image Analysis using FIJI/ImageJ
24-26th of October 2016
CLOSED
ETH Zurich Hönggerberg
The training runs over three consecutive afternoons (1pm - 6pm) and covers the basics of Image Analysis, with a focus on bio-images. The course contains mostly practical hands on sessions with FIJI/ImageJ (with a bit of theory). The course is open to all ETH members or external users upon prior agreement. 80CHF fee is used for printed materials and coffee during breaks. Please apply via online application.
Course credit: 1 ECTS, for PhD students.
Practical course in advanced 3D microscopy
CLOSED
05.09-16.09.2016
ETH Zurich, University of Zurich, and University Hospital Zurich.
The school is dedicated to teaching the basics alongside a wide context of biomedical usage and application that is vital to understand recent advances and current challenges in biological and medical imaging. A wide range of cutting-edge techniques and analyses are intensively discussed to facilitate optimal understanding in this field. To arrive at this goal, leading speakers are consistently holding lectures about the latest developments in their field and likewise give practical sessions to embed the information recently given. Opportunities to network with industry key individuals who are the „movers and shakers“ in their respective fields of influence are afforded to the student.
Excellent MSc and PhD students as well as scientists having a backround in life sciences, computer science, engineering, mathematics and physics are particularly encouraged to apply immediately because of limited qualifying constraints.
Bioimage Analysis Training Schools
CLOSED
NEUBIAS: Network of European Bioimage Analysts
This NeuBIAS training school series is based on the highly successful BIAS courses (@ EMBL) and EuBIAS symposia courses, and will be provided by NeuBIAS faculty and new invited trainers. The program will include computational methods and tools for analysing images of molecules, cells and tissues, tailored for Staff Scientists working in, or in association with, microscopy/bioimaging facilities, who can then provide support and training for researchers who have an immediate need to deploy image analysis in their research. For this, the course extends from basic principles to the actual implementation of workflows using scripting.
Cell Biology and infection: Digital Image Processing/Analysis Tools for Quantitative Light Microscopy Imaging
04.07-08.07.2016
Hellenic Pasteur Institute (HPI), Athens, Greece.
A 5-day practical course, scheduled to take place at the Hellenic Pasteur Institute, Athens Greece, in July 4-8, 2016, with objective to provide theoretical and practical training in basic and advanced concepts and methods of Bioimage informatics.
The course Program will include:- Lectures focused on concepts and methodology for Processing/Analysis of Light Microscopy Digital data (images and videos) and selected topics of Cell Biology and Biology of Pathogen Host interaction.
- Computer practical sessions on the quantitative analysis of microscopy image datasets using the open source software Icy and ImageJ/Fiji
- Informal discussions and tutorials with experts in the field
3rd Copenhagen Image Processing PhD Course
CLOSED
20.06-24.06.2016
University of Copenhagen, Department of Biomedical Sciences, Denmark
The course is suitable not only for beginners in image analysis with no experience, but also for those who want to extend their knowledge of basic principles and more specialised tools. The course is mostly based on interactive lectures given by renowned scientists, where the students will perform themselves all processing and analysis tasks in parallel with the lecturers.
BIAS2016: Bio-Image Data Analysis 2016
(EMBL Master Course)
Application deadline: 2016-04-15
5 - 11 June 2016
EMBL Heidelberg, Germany
This course will focus on computational methods for analysing images of single molecules, cells and tissues, to boost the learning process of participants who have an immediate need to deploy image analysis in their own research. For this, the course extends from basic principles to the actual implementation of workflows using scripting. By increasing their image analysis literacy, participants will greatly enhance the scope, creativity and achievements of their research projects. Expert knowledge will be gathered to create a world-leading course for image analysis in the field of biology
Introduction to Image Analysis using FIJI/ImageJ
20-22nd of April 2016
ETH Zurich Hönggerberg
The training runs over three consecutive afternoons and covers the basics of Image Analysis, with a focus on bio-images. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The course is open to all ETH members or external users upon prior agreement. 80CHF fee is used for printed materials and coffee during breaks. Please apply via online application.
Course credit: 1 ECTS, for PhD students.
Workshop: Image analysis of slide scanner data using Definiens Tissue Studio
REGISTRATION DEADLINE: 12-noon 15th of January 2016.
Feb 16th and 17th, 2016, all-day
ETH Zurich.
Analysing slide scanner data is not an easy task. Even the first step which is the data management and previewing is difficult due to the limits of current state-of-the-art Open Source software e.g. ImageJ/Fiji. To solve this problem, ScopeM offers its users access to Definiens software. The software is one of the most powerful image analysis software available for histology image data sets and is able to read the native 3D-Histech file format. We are now happy to announce a 2-day workshop to image analysis using one of the Definiens products: Tissue Studio. Please apply via link provided above.
Workshop: Analysis and quantification of 4D live-imaged confocal datasets using MorphoGraphX platform
by Anne-Lise Routier-Kierzkowska (Max Planck Institute, Cologne).
09:00-13 HPZ D7.2 Thu. 11th of Feb.
10:00-18 HPZ D7.2 Fri. 12th of Feb.
Scopem is offering to its users a workshop on 4D confocal datasets treatment using the platform called MorphoGraphX. MorphoGraphX [1,2] is an open source application for the visualization and analysis of 4D biological datasets. Developed by researchers, it is primarily used for the analysis and quantification of 4D live-imaged confocal data. Other types of 3D images, such as serial block-face SEM or MRI can also be analyzed in MorphoGraphX. The software is used in various groups to investigate growth, mechanics, gene expression and protein localization at the cellular level, in both plant and animal systems. At this date, 17 works using MorphoGraphX have already been published in prominent journals. In this seminar we will present examples of biological applications of MorphoGraphX, e.g. extracting growth directions or analyzing microtubule orientation. We will also show how plugins can be integrated into the MorphoGraphX platform, for example to extract modeling templates from real data for chemical or mechanical simulations. Over the two days workshop you will have the opportunity to learn the basics of MorphoGraphX and how to use it on your own data.
10th Microscopy Winter School 2016 @ETH Zurich
17-22.01.2016
University of Zurich and ETH Zurich.
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique. Students will be able to apply this technique to their own present and future projects. Practical work consists of eight (8) different modules covering a specific topic each.
Introduction to Image Analysis using FIJI/ImageJ
8-10.12.2015
ETH Zurich Hönggerberg
The training runs over three consecutive afternoons and covers the basics of Image Analysis, with a focus on bio-images. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The course is open to all ETH members or external users upon prior agreement. 75CHF fee is used for printed materials and coffee during breaks. Please apply via online application.
Course credit: 1 ECTS, for PhD students.
EXCITE Summer School 2015:
9th Zurich Summer School on Multiscale Biomedical Imaging
Application deadline: 27.04.2015.
Application: http://www.excite.ethz.ch/education/summer-school/2015/Application.html
31.08-11.09.2015
ETH Zurich, University of Zurich, and University Hospital Zurich.
This course will focus on computational methods for analysing images of single molecules, cells and tissues, to boost the learning process of participants who have an immediate need to deploy image analysis in their own research. For this, the course extends from basic principles to the actual implementation of workflows using scripting. By increasing their image analysis literacy, participants will greatly enhance the scope, creativity and achievements of their research projects. Expert knowledge will be gathered to create a world-leading course for image analysis in the field of biology
2nd Image Processing and Analysis PhD Course
CLOSED
22-26.06.2015, Copenhagen
The course is suitable not only for beginners in image analysis with no experience, but also for those who want to extend their knowledge of basic principles and more specialised tools. The course is mostly based on interactive lectures given by renowned scientists, where the students will perform themselves all processing and analysis tasks in parallel with the lecturers.
BIAS2015: Bioimage Data Analysis
(EMBL Master Course)
Application deadline: Thursday 16 April 2015
Abstract deadline: Thursday 16 April 20157-13.06.2015.
EMBL Heidelberg, Germany
Since the time of Robert Hooke, images observed through microscopes have played a central role in biological investigations. The integration of microscopy with computational science in recent years has dramatically increased its value: images are now considered as multidimensional numerical data, which can be processed and analysed quantitatively using computers. For treating images as measured values knowledge on the nature of digital images, processing protocols and algorithms is required. Skills for using software tools should be accompanied with knowledge of what they are doing. However, methods for extracting quantitative data from micrographs has often been missing in the education of biologists.
This course will focus on computational methods for analysing images of single molecules, cells and tissues, to boost the learning process of participants who have an immediate need to deploy image analysis in their own research. For this, the course extends from basic principles to the actual implementation of workflows using scripting. By increasing their image analysis literacy, participants will greatly enhance the scope, creativity and achievements of their research projects. Expert knowledge will be gathered to create a world-leading course for image analysis in the field of biology
7th Course on Optical Microscopy Imaging for Biosciences
Application link: https://www.ibmc.up.pt/micro15/registration
20-24.04.2015, Porto, Portugal
The 7th Course on Optical Microscopy Imaging for Biosciences is a comprehensive and intensive course to researchers looking for the basis of modern light microscopy. The course explores the principles of optical microscopy including: theory of image formation; the anatomy of an optical microscope; light transmission contrast techniques, fluorescence wide-field and confocal microscopy; digital imaging and quantitative image analysis; live cell imaging; advanced applications for molecular analysis; high-content analysis; super-resolution microscopy.
Introduction to Image Analysis using FIJI/ImageJ
DEADLINE: 1st of May 2015
75 CHF, 10-12 seats.CLOSED
26-28.05.2015, ETH Zurich Hönggerberg
The training runs over three consecutive afternoons and covers the basics of Image Analysis, with a focus on bio-images. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The course is open to all ETH members or external users upon prior agreement. 75CHF fee is used for printed materials and coffee during breaks. Please apply via online application.
Course credit: 1 ECTS, for PhD students.
9th Winter School in Advanced 3D Microscopy 2015
18-24.01.2015
University of Zurich and ETH Zurich.
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique. Students will be able to apply this technique to their own present and future projects. Practical work consists of ten (9) different modules covering a specific topic each (please see description below). Each practical module lasts throughout the whole course. Students will participate in one practical module only. For the practical training the students will use the most state-of-the-art instrumentation. Apart from practical modules, theoretical joined sessions will cover basic knowledge about all the different techniques for all students.
An Industry Day, Wednesday January 21st will provide an overview to the latest developments in instrumentation and techniques currently on the market. Different companies and suppliers in the field of light and electron microscopy will be invited to this event.
Introduction to Image Analysis using FIJI/ImageJ
CLOSED
75 CHF, 10-12 seats. Online application link: http://goo.gl/VNYbrz
24-26.11.2014, ETH Zurich Hönggerberg
The training runs over three consecutive afternoons and covers the basics of Image Analysis, with a focus on bio-images. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The course is open to all ETH members or external users upon prior agreement. 75CHF fee is used for printed materials and coffee during breaks. Online application is available here.
Course credit: 1 ECTS, for PhD students.
8th Zurich Summer School on Biomedical Imaging
CLOSED
1-12.09.2014, ETH Zurich, Switzerland
The school is dedicated to teaching the basics and wider context necessary to understand recent advances and current challenges in biological and medical imaging. Cutting-edge techniques using a wide range of image-formation mechanisms — including magnetic resonance imaging, positron emission tomography, infrared and optical microscopy, electron microscopy and X-ray imaging — will be discussed, with a focus on multimodal and multiscale imaging methods, together with supporting technologies such as computer-aided image analysis and modeling will be discussed.
Practical course in advanced 3D microscopy
CLOSED
12-24.01.2014, ETH Zurich Hönggerberg
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique.
BIAS2014: Bioimage Data Analysis
CLOSED
500 EUR for ACADEMIA, Registration deadline: 25.02.2014
EMBL Heidelberg, Germany
This course will focus on computational methods for analysing images of single molecules, cells and tissues, to boost the learning process of participants who have an immediate need to deploy image analysis in their own research.
ScopeM-IDA is involved in the teaching of this course.Introduction to Image Analysis using FIJI/ImageJ
CLOSED
7-9.04.2014, ETH Zurich Hönggerberg
The training takes three days and covers the basic topics of Image Analysis. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The training is open to all ETH members or external users upon prior agreement. Course credit: 1 ECTS, for PhD students.
Apply by emailing Simon Nørrelykke.Introduction to Image Analysis using FIJI/ImageJ
CLOSED
24-26.03.2014, ETH Zurich Hönggerberg
The training takes three days and covers the basic topics of Image Analysis. The course contains both lectures and practical hands on sessions with FIJI/ImageJ. The training is open to all ETH members or external users upon prior agreement. Course credit: 1 ECTS, for PhD students.
Apply by emailing Simon Nørrelykke.Introduction to IMARIS
CLOSED
13-14.02.2014, ETH Zurich Hönggerberg
The training takes two days and covers the basic functionality of IMARIS and contains both lectures and practical hands on sessions. It is open to all ETH members or external users upon prior agreement.
Apply by emailing Simon Nørrelykke.
Practical course in advanced 3D microscopy
CLOSED
12-24.01.2014, ETH Zurich Hönggerberg
This advanced microscopy course is intended for PhD students and post-graduates with prior experience in microscopy. The goal of the course is to teach and train fundamental knowledge and skills in a specific microscopic technique.
WHO ARE WE?
Core team
Benoit Dehapiot, Ph.D.
Image Analysis
Benoit is a staff scientist with a PhD in Life Sciences, specialising in image data analysis. His expertise includes standard image processing approaches, object segmentation and tracking, as well as advanced data analysis and visualisation techniques. Here at ScopeM, Benoit can help you with your image/data analysis tasks and give courses on these topics.
Area of expertise:
- Image processing
- Data analysis/visualization
- Deep-learning
- Teaching
More information:
Szymon Stoma, Ph.D.
Lead Image Analysis
Szymon is senior staff scientist, holding a Computer Science Ph.D. At ScopeM he leads Image Analysis, teaches and manages IT projects & collaborations. He is passionate about knowledge transfer and startups. CEO of MicroscopeIT - a subsidiary of Tooploox - Custom Software Development Company where he holds a position of Head of R&D.
Area of expertise:
- Image analysis
- Teaching
- Software development
- HPC
- Startups / Tech. transfer
More information:
Andrzej Rzepiela, Ph.D.
Lead Data Analysis
Andrzej is senior staff scientist interested in questions that can be approached with statistical modeling. In the past he focused on numerical simulations & mathematical models in the area of systems biology. Andrzej leads Data Analysis, manages projects and expertise in data analysis and data management.
Area of expertise:
- Data analysis
- Data management
- Machine learning
- Bioinformatics
More information:Members

Joanna Kaczmar
Visiting Scholar
Joanna is a Machine Learning researcher, fascinated by the potential of artificial intelligence in application to molecular chemistry. She holds a Master’s degree in Computer Science and has extensive experience in applying deep learning in commercial context. In IDA she is researching the use of deep learning models in the field of drug discovery.
Area of expertise:
- Machine Learning
- Software development
Alumni

Javier Montoya Zegarra, Ph.D.
Postdoc
Javier is a Computer Vision expert who wrote his PhD thesis about reconstruction of road networks from aerial images. He was working on image analysis projects from the groups of Prof. Cornelia Halin and Prof. Michael Detmar. He moved to join the group of Prof. Sebastian Kozerke.

Alvaro Gomariz
PhD candidate
Alvaro is a biomedical engineer who wrote his Master's Thesis with ScopeM. Alvaro investigates the intercellular interactions within mouse bone marrow tissue based on 3D microscopy images. After copleting a PhD with Prof. Cesar Nombela-Arrieta and Prof. Orcun Goksel he joined Novartis.

Gabriela Evrova
Master Student
Gabriela is pursuing her education in Computer Science. She earned a CS Bachelor degree at EPFL, now she joined the Master program in ETH Zurich. Gabriela supports our developments in R&D projects and helps us to deal with a daily IDA business. Currently Gabriela joined Magic Leap.

Nelly Hajizadeh, Ph.D.
Post doc
Nelly is a postdoc with a Ph.D. in Biostatistics/Biophysics and an interested in methods development for biological data-analysis. She has experience with developing statistical data-analysis methods for biophysical and structural biological data. Her aim in the context of IDA is to apply deep learning to improve the image analysis pipeline for electron microscopy data. Currently Nelly joined Novartis.

Krzysztof Lis
PhD cand.
Krzysztof is a Master Student interested in Computer Vision. His current interests are centered around visual perception for autonomous cars: i) detecting unusual and anomalous objects, ii) semantic segmentation, iii) jointly perceiving semantic and 3D information about the scene. Currently PhD student at the EPFL Computer Vision Lab of Pascal Fua.

Beda Rutishauser
Bachelor Student
Beda is a Bachelor student in Computer Science with knowledge in the field of physics and math. At IDA, he is improving his skills in handling data and the application of machine learning and image analysis models. His aim is to put his skills in these fields on the same solid pillars as he did as a former polymechnician in applied mechanics. Currently in ABB.

Ishaant Agarwal
Master Student
Ishaant is a Master's student interested in the intersection of artificial intelligence and neuroscience. He is double majoring in Physics and Electronics Engineering and has previous experience in computational and behavioral neuroscience, as well as image analysis. At IDA he is looking to develop neural network models to denoise 3D images like cryo-EM and MRIs, for his honors thesis. Currently in Oracle.

Manjunatha Bairi
Master Student
Manjunatha is a Master's student interested in the application of deep learning to image processing tasks involving segmentation and super resolutions. He is in the process of completing his integrated masters in Chemistry and Electronics and has worked on facial expression transfers and modeling of reinforcement learning environments in the past. At IDA he is researching the impact of synthetic data in the training of nuclei segmentation models.

Simon Flyvbjerg Nørrelykke, Ph.D.
Staff Scientist
Simon holds a Ph.D. in Physics from Copenhagen University and has since worked in Florence, Dresden, and Princeton. His past research spans econo-physics, optical tweezers, single-molecule biophysics, and cell migration. At ScopeM he taught and practiced image processing and data analysis and headed the IDA group from 2012--2022.
Starting September 2022 Simon can be found in Boston at Harvard Medical School.
Recent publications (2016+)
Please check a complete list based on PUBMED search
Emmanouela Rantsiou, Franziska Oschmann, Lukas von Ziegler, Thomas Wüst, Andrzej J. Rzepiela, Szymon Stoma
F1000Research 2025

Joel Lüthi, Marvin Albert, Liviu Anita, Kola Babalola, Davis Bennett, John A. Bogovic, Lorenzo Cerrone, Rémy Dornier, Jan Eglinger, Vera Galinova, Reto Gerber, Oane Gros, Stefan Hahmann, Max Hess, Ruth Hornbachner, Dmytro Horyslavets, Rachael Huxford, Daniel Krentzel, Tong Li, Luca Marconato, Matthew McCormick, Franziska Moos, Filip Mroz, Bugra Özdemir, Benjamin Pavie, Eric Perlman, Maximilian Schulz, Leonardo Schwarz, Hannes M. Spitz, David Stansby, Fabio Steffen, Szymon Stoma, Flurin Sturzenegger, Wouter-Michiel Vierdag, Jonas Windhager, Kevin A. Yamauchi, Igor Zubarev, Josh Moore, Norman Rzepa, Christian Tischer, Vladimir Ulman, Virginie Uhlmann
BioHackrXiv 2025

Ishaant Agarwal, Joanna Kaczmar-Michalska, Simon F. Nørrelykke and Andrzej J. Rzepiela
IUCrJ, 2024

Joanna Kaczmar-Michalska & Andrzej J. Rzepiela
Lecture Notes in Networks and Systems, 2024

Leonard Krupnik, Jonathan Avaro, Marianne Liebi, Neda Iranpour Anaraki, Joachim Kohlbrecher, Alla Sologubenko, Stephan Handschin, Andrzej J. Rzepiela, Christian Appel, Tiberiu Totu, Clement E. Blanchet, Amy E. Barton Alston, Reinaldo Digigow , Erik Philipp, Beat Flühmann, Bruno F.B. Silva, Antonia Neels, Peter Wick
Journal of Controlled Release, 2024

Mechanisms of Sarcomere Assembly in Muscle Cells Inferred from Sequential Ordering of Myofibril Components
F Kolley, C Sidor, B Dehapiot, F Schnorrer, BM Friedrich
PRX Life, 2024

Mechanical regulation of substrate adhesion and de-adhesion drives a cell-contractile wave during Drosophila tissue morphogenesis
C Collinet, A Bailles, B Dehapiot, T Lecuit
Developmental Cell, 2024

Moreau A, Yaya F, Lu H, Surendranath A, Charrier A, Dehapiot B, Helfer E, Viallat A, Peng Z.
Proc Natl Acad Sci USA, 2023

Mercedes Thieme, Anton Hochmuth, Theresa Elisabeth Ilse, Jose A Cuesta-Seijo, Szymon Stoma, Roger Meier, Simon Flyvbjerg Nørrelykke, Pai Rosager Pedas, Ilka Braumann, Samuel C Zeeman
Carbohydrate Polymers, 2022

Paolo Zanoni, Grigorios Panteloglou, Alaa Othman, Joel T Haas, Roger Meier, Antoine Rimbert, Marta Futema, Yara Abou-Khalil, Simon Flyvbjerg Nørrelykke, Andrzej Rzepiela, Szymon Stoma, Michael Stebler, Freerk van Dijk, Melinde Wijers, Justina C Wolters, Nawar Dalila, Nicolette Huijkman, Marieke Smit, Antonio Gallo, Valerie Carreau, Anne Philippi, Jean-Pierre Rabès, Catherine Boileau, Michele Visentin, Luisa Vonghia, Jonas Weyler, Sven Francque, An Verrijken, Ann Verhaegen, Luc F Van Gaal, Adriaan van der Graaf, Belle V van Rosmalen, Jérôme Robert, Srividya Velagapudi, Mustafa Yalcinkaya, Michaela Keel, Silvija Radosavljevic, Andreas Geier, Anne Tybjærg-Hansen, Mathilde Varret, Lucia Rohrer, Steve E Humphries, Bart Staels, Bart van de Sluis, Jan Albert Kuivenhoven, and Arnold Von Eckardstein

Bolck, Hella A. and Przetocka, Sara and Meier, Roger and von Aesch, Christine and Zurfluh, Christina and Hänggi, Kay and Spegg, Vincent and Altmeyer, Matthias and Stebler, Michael and Nørrelykke, Simon F. and Horvath, Peter and Sartori, Alessandro A. and Porro, Antonio
Cells, 2022
Kota Miura and Simon F Nørrelykke
The EMBO Journal 2022

P. Zanoni, G. Panteloglou, A. Othman, J.T. Haas, R. Meier, A. Rimbert, M. Futema, Y. Abou Khalil, S.F. Nørrelykke, A.J. Rzepiela, S. Stoma, M. Stebler, F. Van Dijk, M. Wijers, J.C. Wolters, N. Dalila, N. Huijkman, M. Smit, A. Gallo, V. Carreau, A. Philippi, J.-P. Rabès, C. Boileau, M. Visentin, L. Vonghia, J. Weyler, S.M. Francque, A. Verrijken, A. Verhaegen, L. Van Gaal, A. Van Der Graaf, B.V. Van Rosmalen, S. Velagapudi, M. Yalcinkaya, M. Keel, S. Radosavljevic, A. Geier, A. Tybjærg-Hansen, M. Varret, L. Rohrer, S.E. Humphries, B. Staels, B. Van De Sluis, J.A. Kuivenhoven, A. Von Eckardstein
Atherosclerosis, 2020

Beth A. Cimini, Simon F. Nørrelykke, Marion Louveaux, Nataša Sladoje, Perrine Paul-Gilloteaux, Julien Colombelli, Kota Miura
F1000Research, 2020
Edited by Kota Miura and Nataša Slodoje.
Springer, 2020
https://doi.org/10.1007/978-3-030-22386-1
Supplementary material (code and images) on GitHub:
Samia B Bachmann, Denise Gsponer, Javier A Montoya-Zegarra, Martin Schneider, Felix Scholkmann, Carlotta Tacconi, Simon F Nørrelykke, Steven T Proulx, Michael Detmar
Cell Reports, 2019

Stefanie S Schmieder, Claire E Stanley, Andrzej Rzepiela, Dirk van Swaay, Jerica Sabotič, Simon F Nørrelykke, Markus Aebi, Markus Künzler

Catharina D Commerford, Lothar C Dieterich, Yuliang He, Tanja Hell, Javier A Montoya-Zegarra, Simon F Noerrelykke, Erica Russo, Martin Röcken, Michael Detmar
Cell Reports, 2018

Javier A Montoya-Zegarra, Erica Russo, Peter Runge, Maria Jadhav, Ann-Helen Willrodt, Szymon Stoma, Simon F Nørrelykke, Michael Detmar, Cornelia Halin
Angiogenesi, 2018

Alvaro Gomariz, Patrick M Helbling, Stephan Isringhausen, Ute Suessbier, Anton Becker, Andreas Boss, Takashi Nagasawa, Grégory Paul, Orcun Goksel, Gábor Székely, Szymon Stoma, Simon F Nørrelykke, Markus G Manz, César Nombela-Arrieta
Nature Communications, 2018.

Srividya Velagapudi, Mustafa Yalcinkaya, Antonio Piemontese, Roger Meier, Simon Flyvbjerg Nørrelykke, Damir Perisa, Andrzej Rzepiela, Michael Stebler, Szymon Stoma, Paolo Zanoni, Lucia Rohrer, Arnold von Eckardstein
Arteriosclerosis, Thrombosis, and Vascular Biology, 2017

Lehmann S, Te Boekhorst V, Odenthal J, Bianchi R, van Helvert S, Ikenberg K, Ilina O, Stoma S, Xandry J, Jiang L, Grenman R, Rudin M, Friedl P.
Current Biology, 2017

Versari C, Stoma S, Batmanov K, Llamosi A, Mroz F, Kaczmarek A, Deyell M, Lhoussaine C, Hersen P, Batt G.
Journal of Royal Society Interface, 2017


Lehmann S, Perera R, Grimm HP, Sam J, Colombetti S, Fauti T, Fahrni L, Schaller T, Freimoser-Grundschober A, Zielonka J, Stoma S, Rudin M, Klein C, Umana P, Gerdes C, Bacac M.
Clinical Cancer Research, 2016
CONTACT
We are located in:
Scientific Center for Optical and Electron Microscopy (ScopeM, HPZ G34/35)
John-von-Neumann-Weg 9, 8093 Zurich
or contact us through any of these channels!

Phone
+41 766 390 449
Design: Szymon Stoma
Copyright 2013-2024, ETH